This is an old revision of the document!
Tools for Molecular Sistematics
Sequence submission to the NCBI/GenBank
Here I will provide some information on how to submit sequences to NCBI/GenBank using Sequin. It is the goal of this tutorial to make sure we standardize the information we associate with the sequences. This is important because not all the sequences deposited at NCBI for our group provide the same information (i.e, host data, voucher; among others) and even if they do, they are not in a standard format - that is, under the same source modifier (see bellow).
The information associated with sequences in the NCBI/GenBank is done by Source Modifier. The list of source modifiers provided by the NCBI/GenBank is long and among those we think that we should use at least these: Organism, specimen-voucher, isolate, dev_stage host, collected-by, collection-date, country, lat_lon, and note.
Table of modifiers
The first thing you have to build is a table with the following content:
| SeqId | Organism | specimen-voucher | isolate | dev_stage | host | collected-by | collection-date | country | lat_lon | note |
|---|---|---|---|---|---|---|---|---|---|---|
| Pararhin_EC_14.1 | Pararhinebothroides hobergi | MZUSP 7754 | EC-14.1 | adults | Urobatis tumbesensis | J. N. Caira & F. Marques | 24-May-2014 | Ecuador | S 2.19967, W 80.9780 | Host field number EC-14 |
| Pararhin_EC_56.1 | Pararhinebothroides hobergi | MZUSP 7755 | EC-56.1 | adults | Urobatis tumbesensis | J. N. Caira & F. Marques | 3-Jun-2014 | Ecuador | S 2.19967, W 80.9780 | Host field number EC-56 |
| Pararhin_EC_56.2 | Pararhinebothroides hobergi | MZUSP 7756 | EC-56.2 | adults | Urobatis tumbesensis | J. N. Caira & F. Marques | 3-Jun-2014 | Ecuador | S 2.19967, W 80.9780 | Host field number EC-56 |
| Pararhin_EC_56.3 | Pararhinebothroides hobergi | MZUSP 7757 | EC-56.3 | adults | Urobatis tumbesensis | J. N. Caira & F. Marques | 3-Jun-2014 | Ecuador | S 2.19967, W 80.9780 | Host field number EC-56 |
Here, the first row contains the source modifiers. The Sequence ID (SeqId) is the name of your sequence in the FASTA file. Thus remember, it is mandatory that the first column name matches the name of your sequences in the FASTA file. The other columns are pretty intuitive, but I should comment some modifiers. The isolate should receive your molecular code. The date of collection (date) has to be in the format: DD-month-YYYY (i.e., “24-May-2014”). The latitude and longitude data (lat_lon) should be inserted in a “semi-decimal” format (i.e., “S 2.19967, W 80.9780”). I say “semi-decimal” format because in LAT/LONG in an decimal format uses +/- for N/S and E/W, respectively. If your georeference data is in Degrees, Minutes, Seconds format, you can covert using Montana State University webpage. Finally, the host field number should be inserted under the source modifiers note, as in “Host field number XXXX”.
You can build this table in any spreadsheet application, but you will have to save that as a TAB delimited text file. You can download the table above in a TAB delimited format here.
Running Sequin
Once you have your table built, you can run Sequin. I will go through all the steps of Sequin using the sequence FASTA file below, which I will call example.fas. It is important to degap your sequences, that is, you should always submit unaligned nucleotide sequences.
>Pararhin_EC_14.1 ACAGGATGAGCCAGGATTCGCTCGGACACGATGGGCACGAGATTAGGACACAG >Pararhin_EC_56.1 ACAGGATGAGCCAGGATTCGCTCGGACACGATGGGCACGAGATTAGGACACAG >Pararhin_EC_56.2 ACAGGATGAGCCAGGATTCGCTCGGACACGATGGGCACGAGATTAGGACACAG >Pararhin_EC_56.3 ACAGGATGAGCCAGGATTCGCTCGGACACGATGGGCACGAGATTAGGACACAG
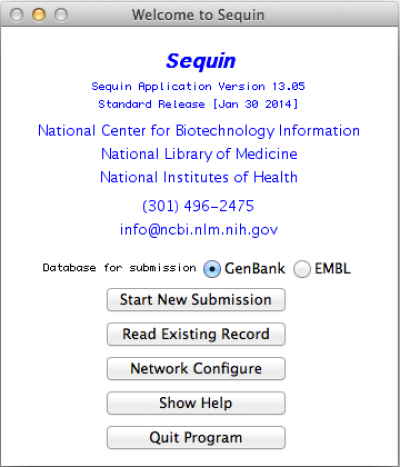
So, as expected, the first thing to do is to open Sequin. You should get the following window:
In this window, you should click on “Start New Submission”. If you have already done one and want to use some of your data from the previous record (i.e., address, affiliation, among other) you can click on “Read Existing Record”. Regardless of your choice, you should get the following window:
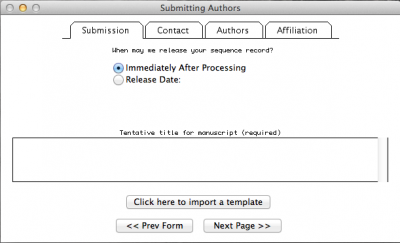
Here you will insert all the information concerning the publication to which your sequences will be related, your contact, authorship, and affiliation information.
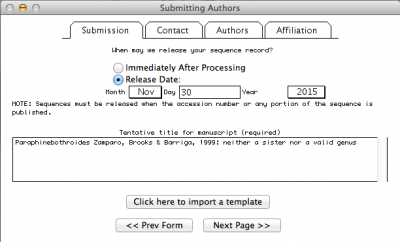
Note: In the window above, I selected the option in which I define the date in which the data should be public. By default, NCBI will give you one year. But you can make it longer or shorter as well as you can change the status of you sequence at any time by contacting the NCBI personal.
This step is pretty intuitive, after filling up all the fields in “Contact”, “Authors”, and “Affiliation” just click on the button “next page”. By doing so, you will obtain:
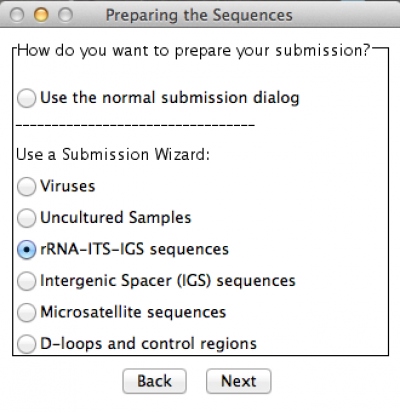
In this example, I am submitting a rDNA 28S sequence. Thus, I chose the option accordingly. Press “Next” and you will get the following window:
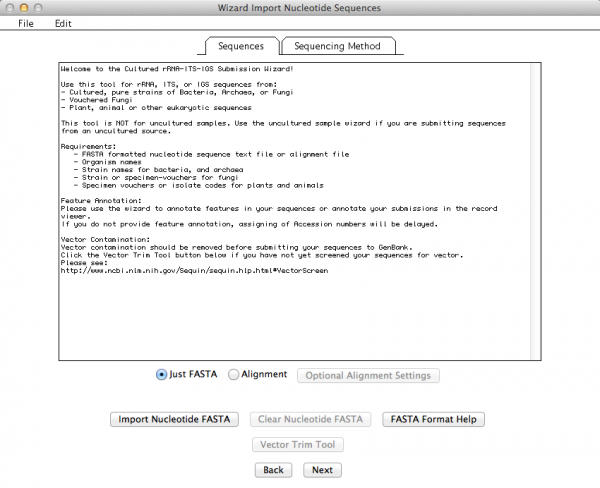
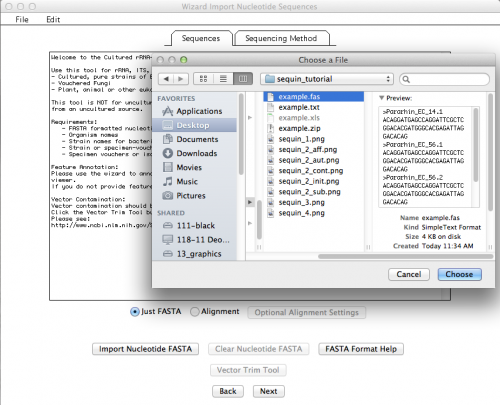
This is the window that allows you to import your External FASTA file. If you click on “Import Nucleotide FASTA”, you will get the option to choose the file you want to import (Remember: your file must have unaligned sequences!):
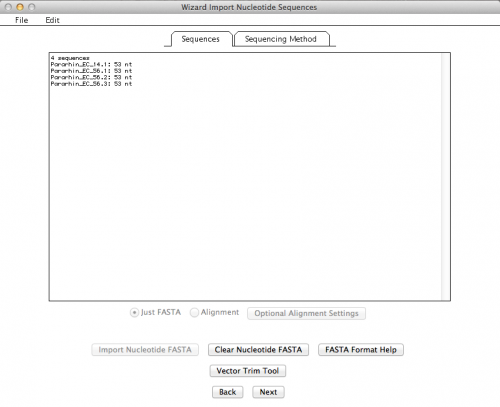
If all went well, you should get a summary of your imported sequences:
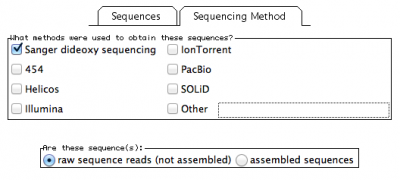
In this window, you will also have to provide the methodology you used to obtain your sequences under the tab “Sequencing Method”:
Here I selected sanger sequencing and indicated the they are “raw sequence reads”.
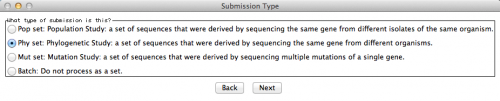
The next window will ask you the type of submission you are about to do. In this particular case, I want to associate the sequences with a phylogenetic study. Therefore, here is the option I selected:
Go to the next window.
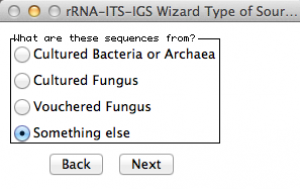
Here, Sequin will ask from which organism you got your sequences. As you can see, there is no option for worms! Thus, I selected “something else”!
Move on to the next:
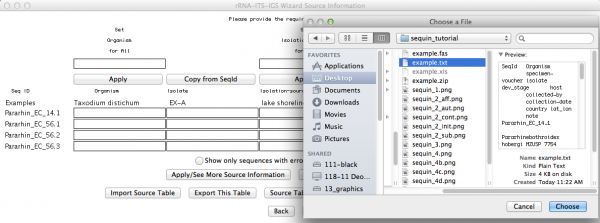
Here is an important step, since you will now use the table with the source modifiers you did as the first part of this tutorial. You should click on “import source table” and select the file. Once accomplished this step, go to the next window.
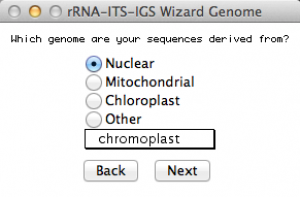
This window will ask you from which genome you obtained your sequences. Since we are submitting sequences of rDNA 28S, I selected Nuclear genome. Once you selected an option, go to the next.
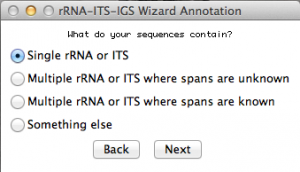
Now you will have to select the type of fragment or genomic region you sequenced. In that case, I selected “single rRNA or ITS”. Move to the next.
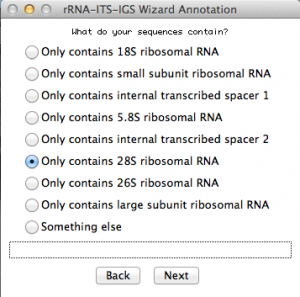
Because I selected “single rRNA or ITS”, this window will ask some more details about my sequences. Select accordingly. In this case, I selected the option for rDNA 28S. Once you are done, go to the next:
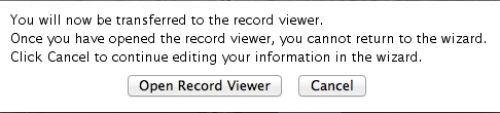
Now you are basically done! You should select “Open Record Viewer”, because you will need to verify whether there are errors in your data and you will have to save the file to be sent to NCBI.
When you press “Open Record Viewer”, if no errors are found and reported, you should get the following:
Note: If you got an error message, please see the section on Error Reports below.
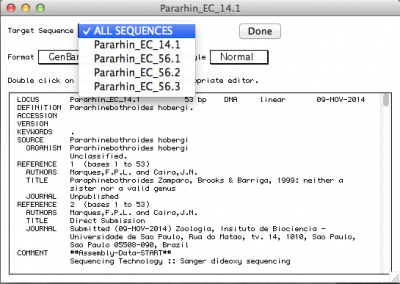
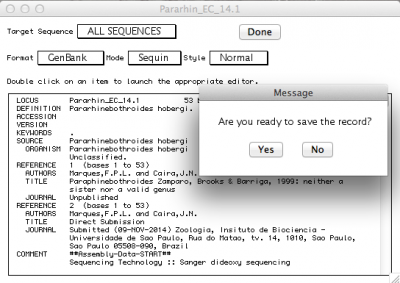
By default, the record viewer shows only the first sequence of your file. You could inspect one by one, to make sure all the information you desired is associated with your sequence. To finally export the file to be submitted to NCBI, you should select “ALL SEQUENCES” in “Target sequences” and click on “Done”.
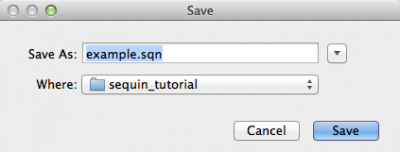
Sequin will then ask if you want to save the record. Press “Yes”, and you will be presented with the following window:
Here you should give a name to the file and press “Save”.
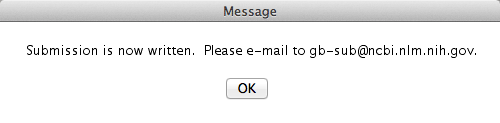
Congratulations! You are done! Well, in part. Sequin finished its job, which was to generate a file compatible to the input files NCBI receive the sequences. After you saved the file, you will get the following warning message:
Pay attention to that! Here is the email to which you should send the file Sequin just generated. So, I guess, your job will be done once you email the file!
Error Reports in Sequin
According to NCBI, “[t]here are four types of error messages, Info, Warning, Error, and Reject. Info is the least severe, and Reject is the most severe. You may submit the record even if it does contain errors. However, we encourage you to fix as many problems as possible. Note that some messages may be merely suggestions, not discrepancies. A possible Warning message is that a splice site does not match the consensus. This may be a legitimate result, but you may wish to recheck the sequence. A possible Error message is that the conceptual translation of the sequence that you supplied does not encode an open reading frame. In this case, you should check that you translated the sequence in the correct reading frame. A possible Reject message is that you neglected to include the name of the organism from which the sequence was derived. The name of the organism is absolutely required for a database entry. If Sequin does not detect any problems with the format of your record, you will see a message that “Validation test succeeded”.
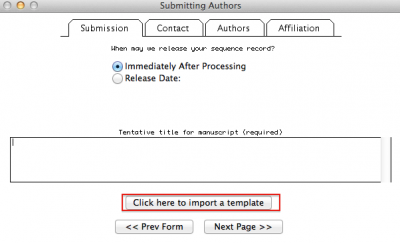
If you got an error message and was able to identify the error, you can reopen Sequin, start a new submission, but instead of following all the steps we discuss above, you select “Click here to import a template”. See figure below:
After importing the data, follow the steps of the program, which were discussed above. Good luck!
Tree editing and manipulation
Below I will give you some tips on how to deal with file manipulation during your phylogenetic analysis in order to avoid a lot of editing of file. As a rule of thumb, I like to keep the name of my terminals short and without characters that are bound to give you troubles as you migrate files from one application to the other (i.e., ”-“, ”&“, ”#“, among others). Underscore (i.e., “_”) is your only option most of the time.
Consider the following example of data file:
>Potamotrygonodestus_fitzgeraldae_amazon_PA03-53.1 ATTTACTATCATTCACATATCAAGAATGATGACAGATGAGCATGAC... >Potamotrygonodestus_fitzgeraldae_amazon_PA03-53.2 ATTTATTAACATACACATATCAAGAATGATGACAGATGAGCATGAC... >Potamotrygonodestus_marajoara_amazon_PA03-13.1 ATTTATTAACATACACATATCAAGAATGATGACAGATGAGCATGAC... >Potamotrygonodestus_marajoara_amazon_PA03-13.2 ATTTATTAACATACACATATCAAGAATGATGATTTTTGAGCATTTC... ...
This format, encompasses the name of the species, its distribution and a code for the specimen sequenced. Some programs, and TNT is an example, would truncate names like this to 32 characters. Therefore, these programs would consider each of these taxon names as:
>Potamotrygonodestus_fitzgeraldae ATTTACTATCATTCACATATCAAGAATGATGACAGATGAGCATGAC... >Potamotrygonodestus_fitzgeraldae ATTTATTAACATACACATATCAAGAATGATGACAGATGAGCATGAC... >Potamotrygonodestus_marajoara_am ATTTATTAACATACACATATCAAGAATGATGACAGATGAGCATGAC... >Potamotrygonodestus_marajoara_am ATTTATTAACATACACATATCAAGAATGATGATTTTTGAGCATTTC ...
Note that the two initial taxa would have the same name, the same is true for the last two, and that would be enough impede you to analyze this data.
I am aware we give names that would allow us to recognize the taxa as fast as possible. That I will try to convince you is that you can keep code names in your input files and use a simple tool to make the translations required whenever you need to visually examine the result of your analysis.
Here is how I would deal with that:
Usually, I use a code for each specimen. If I am dealing with hundreds of terminal, I use few letters and at least three digits. For instance, for the example above I would have something like:
>Pot001 ATTTACTATCATTCACATATCAAGAATGATGACAGATGAGCATGAC... >Pot002 ATTTATTAACATACACATATCAAGAATGATGACAGATGAGCATGAC... >Pot003 ATTTATTAACATACACATATCAAGAATGATGACAGATGAGCATGAC... >Pot004 ATTTATTAACATACACATATCAAGAATGATGATTTTTGAGCATTTC... ...
Those taxon names will never give you a hard time in any application I am aware of! But of course, you need to keep track of what those codes means and most of the time, we do that by having a spreadsheet that document all our specimens or terminals. Let us then consider a two column table:
| Code | Specimen |
|---|---|
| Pot001 | Potamotrygonodestus_fitzgeraldae_amazon_PA03-53.1 |
| Pot002 | Potamotrygonodestus_fitzgeraldae_amazon_PA03-53.2 |
| Pot003 | Potamotrygonodestus_marajoara_amazon_PA03-13.1 |
| Pot004 | Potamotrygonodestus_marajoara_amazon_PA03-13.2 |
From this table, you can generate a text file called rename.sed in the following format:
s/Pot001/Potamotrygonodestus_fitzgeraldae_amazon_PA03_53_1/ s/Pot002/Potamotrygonodestus_fitzgeraldae_amazon_PA03_53_2/ s/Pot003/Potamotrygonodestus_marajoara_amazon_PA03_13_1/ s/Pot004/Potamotrygonodestus_marajoara_amazon_PA03_13_2/
This is a file formatted for Sed; a stream editor native to UNIX/LINUX systems, therefore also available in your Mac. Sed is a powerful toll when it comes to make substitutions in text files - remember, most of the files we manipulate during phylogenetic analyses are text files! Note: I did avoid to use ”-“ and ”.“, because depending on what you want to do, those characters will not work!
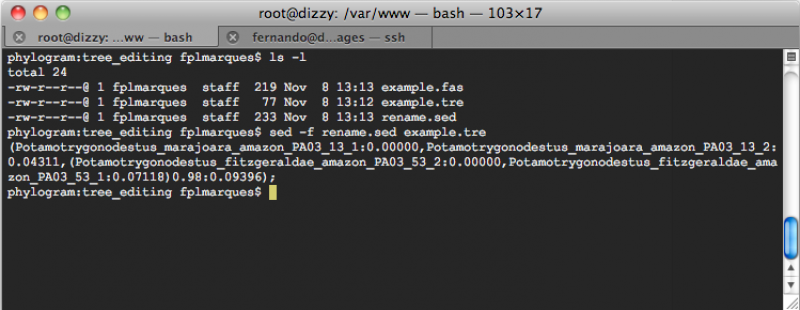
Here it how you can apply this tool. Consider that you for a tree output file, which we will call example.tre, with the following content:
(Pot003:0.00000,Pot004:0.04311,(Pot002:0.00000,Pot001:0.07118)0.98:0.09396);
Now, if you open a terminal window, move to the directory in which your files are, and execute the following command line:
sed -f rename.sed example.tre
You will get:
However, you may desire to redirect the output of the commmand line above to a file so that you can open the file in something like FigTree. In that case, you should execute the following command line:
sed -f rename.sed example.tre > example_renamed.tre
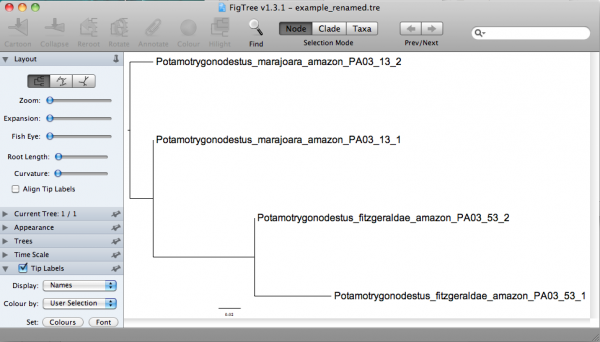
In this command line, the option ”-f“ of sed implied that all rules of substitution are in a file (i.e., rename.sed) and that the file to be translated is example.tre and the output of this sed command line should be redirected (the sign ”>“ is what does it) to the file example_renamed.tre. The content of the file example_renamed.tre should be:
(Potamotrygonodestus_marajoara_amazon_PA03_13_1:0.00000,Potamotrygonodestus_marajoara_amazon_PA03_13_2:0.04311,(Potamotrygonodestus_fitzgeraldae_amazon_PA03_53_2:0.00000,Potamotrygonodestus_fitzgeraldae_amazon_PA03_53_1:0.07118)0.98:0.09396);
Now you should be able to examine this tree in a more informative way. If you open that in FigTree, you would have:
But you can do better, especially if you are looking at a final tree, which would be used to genetate a figure for publication. Considere this substitution file, which we will call rename_2.sed:
s/Pot001/"Potamotrygonodestus fitzgeraldae ex P. motoro PA03-53.1 [Amazon\/AJ51651]"/ s/Pot002/"Potamotrygonodestus fitzgeraldae ex P. motoro PA03-53.2 [Amazon\/AJ51652]"/ s/Pot003/"Potamotrygonodestus marajoara ex Pl. iwamae PA03-13.1 [Amazon\/AJ51653]"/ s/Pot004/"Potamotrygonodestus marajoara ex Pl. iwamae PA03-13.1 [Amazon\/AJ51654]"/
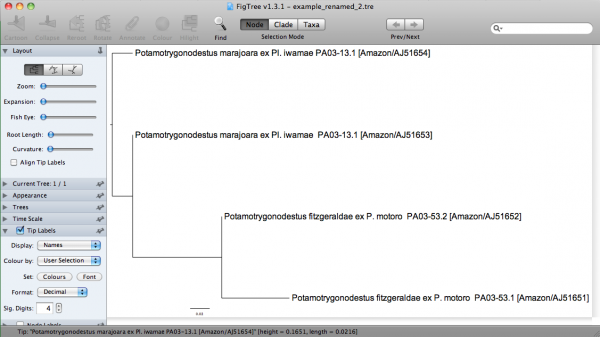
Note that I am using all kinds of characters that are bound to give you a lot of hassle when using in input files. One of the special characters (i.e., ”/“) in “Amazon/AJ51651” requires a back slash, since sed consider ”/“ a special character; hence, “Amazon\/AJ51651”. There I am enclosing them between quotes, because FigTree allows me to do that. If I execute the command line above using this substitution file, I would get:
("Potamotrygonodestus marajoara ex Pl. iwamae PA03-13.1 [Amazon/AJ51653]":0.00000,"Potamotrygonodestus marajoara ex Pl. iwamae PA03-13.1 [Amazon/AJ51654]":0.04311,("Potamotrygonodestus fitzgeraldae ex P. motoro PA03-53.2 [Amazon/AJ51652]":0.00000,"Potamotrygonodestus fitzgeraldae ex P. motoro PA03-53.1 [Amazon/AJ51651]":0.07118)0.98:0.09396);
In FigTree, this tree would be: